Research Article
Molecular study of Apolipoprotein E gene in familiar hypercholesteromic families
Nasir Ali1*, Sajid Naeem2, Suliman Qadir Afridi3, Javed Muhammad4, Umar Bacha5
Adv. life sci., vol. 1, no. 1, pp. 13-17, November 2013
*-Corresponding Author: Nasir Ali (Email: nasirimbb@gmail.com)
Author Affiliations
2-Institute of Industrial Biotechnology, Government College University, Lahore – Pakistan
3-Department of Microbiology, University of Veterinary and Animal Sciences, Lahore – Pakistan
4-University Diagnostic Laboratory, University of Veterinary and Animal Sciences, Lahore – Pakistan
5-Department of Food Science and Human Nutrition, University of Veterinary and Animal Sciences, Lahore – Pakistan
Abstract
Introduction
Methods
Results
Discussion
References
Abstract
Background: Familial hypercholesterolemia (FH) is understood to be one in all the foremost common hereditary disease and critically associated with Coronary heart condition worldwide. FH is taken into account to be caused because of mutations and polymorphisms within the apolipoprotein E (Apo E) cistron. Exaggerated level of density compound protein LDL-C is that the hallmark of this malady.
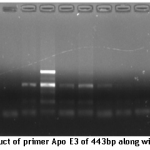
Methodology: Seven hypercholesterolemic families were chosen for this study. Case history was taken and pedigree was created in person by visiting every family. Exon3 and exon4 regions of ApoE cistron were amplified using polymerase chain reaction (PCR). After successful amplification, both citrons were sequenced. Single strand conformation polymorphism (SSCP) results were obtained to support the different pattern of single strand polymorphism of studied samples.
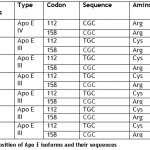
Results: The sequencing results of probands from all the seven families showed that six out of seven have Apo E three isoform whereas one family showed change within the sequence from T to C at 112 sequence position of processed macromolecule resulted in amino acid that represents it as Apo E4 isoform.
Conclusion: Our findings show that Apo E3 is more prevalent than Apo E4 and other isoforms in studied population of Pakistan.
Keywords: Familial hypercholesterolemia, Apolipoprotein E, Single strand conformation polymorphism, Apo E3
Introduction
Familial hypercholesterolemia (FH) is thought to be one in all the inheritable disorders that is said to be caused due to abnormality in metabolism of plasma lipoprotein. FH is usually manifested by an elevated level (LDL)-cholesterol that ends up in several complications like lipid accumulation in vessels of blood provides, premature arterial sclerosis and nice risk of diseases associated with heart [1]. Clinical and molecular characterization of lipid metabolism describes the FH as firs genetic disease [2].
FH is associate degree chromosome attribute, and loosely characterised into 2 classes supported clinical manifestation and phenotypical prevelance of “heterozygous” and “homozygous” form. blood serum LDL-C level is twice and 4 fold higher in heterozygous and homozygous respectively [3]. FH prevalence relating to heterozygous is roughly one in five hundred individuals (10 million world-wide) and therefore the increased serum cholesterol concentration leads to coronary cardiovascular disease [4]. half the people with FH don't survive before the age of sixty attributable to infarction [5]. The homozygous kind of FH is comparatively rare as compared to heterozygous type (1/1000000 live born) and is much more fatal counting on what variety of mutation could occur and results death within the early age of life [6].
Pakistan has additionally a high illness burden of infarction and its risk factors. Coronary artery disease is found to be the underlying reason for infarction within the overwhelming majority from Islamic Republic of Pakistan [7]. It is reported that at the age of 45% of male and 20% of female patients suffers from coronary artery disease (CAD) [2]. The heterozygous variety of FH is treated by numerous ways like regular exercise, intake of assorted medicinal drug medication and fiber made diet.
Methods
Identification and Sampling of FH positive families
Families with a minimum of 2 or additional individual clinically diagnosed with symptom were selected from completely different areas of District Mardan of Khyber Pukhtoon Khuwa and district Lahore of Punjab, Pakistan. Seven families were designated for this study. Case history was taken and pedigree was made personally by visiting every family. Elaborate history was taken from every family to reduce the presence of different abnormalities and environmental causes of symptom. different relatives of the affected family with symptom were conjointly enclosed within the study counting on their willingness and availableness. Informed consent was obtained for taking part within the study.
Blood samples (5 mL) were collected from all the affected people, their traditional siblings, folks to trace the mode of inheritance. The blood samples were collected in 5 mL vacutainer already containing heparin that work as an anticoagulant, the blood samples were kept in ice boxes in real time when their collection was made and stored in -200C before DNA extraction. Pedigrees were drawn on Cyrillic software V.4.0 with the help of data taken from each affected family. At least three or four generation family data as (sibs, cousin marriage, monozygotic twin and sex) was shown by biological symbols. DNA was extracted by in-organic method and stored at -80oC for later use [8].
PCR for Apo E gene polymorphisms
The complete sequence of two primers Apo E3 and Apo E 4 were selected from the National Center of Biotechnology Information (NCBI) website at URL (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi). Specific primers for the selected exons of Apo E were designed using Primer3 software web facility [9]. All the primers were optimized on the BioRad Thermocycler. The conditions were set at which the best results were obtained. Amplification reactions were carried out at temperatures and recipe specific for each primer.
Results
A total of seven families with 2 or additional affected people, clinically diagnosed with hypercholesteremia were known from totally different areas of district Mardan and Lahore of Khyber Pukhtoon Khuwa and Punjab respectively. Proband sample of every family were sequenced using genetic analyzer ABI3130xL. The sequencing results of probands from all the seven families represent that six out of seven have Apo E three isoform whereas one family showed modification within the sequence from T to C at 112 sequence position of processed protein resulted in cystine to arginine that indicate it as Apo E4 isoform. Position of processed amino acid 112 and 158 are 130 and 176 codon respectively in transcript strand of mRNA (Ensembl.org). Our findings show that Apo E3 is more prevalent than other isoforms. Similar findings were also reported by using the restriction analysis [10].
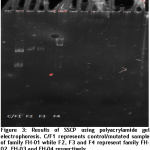
SSCP
Single strand conformation polymorphism (SSCP) results were obtained to support the different pattern of single strand with other samples. The sequenced sample of family FH-01 was considered as standard and a single sample from each family was taken to compare the pattern.
Discussion
In present research, we determined the of Apo E polymorphism frequency in seven families of district Mardan of Khyber Pukhtoon Khuwa and district Lahore of Punjab. The sequencing results of probands from all the seven families represent that six out of seven have Apo E3 isoform while one family showed change in the sequence from T to C at 112 codon position of processed protein resulted in cystine to arginine which indicate it as Apo E4 isoform. Position of processed amino acid 112 and 158 are 130 and 176 codon respectively in transcript strand of mRNA (Ensembl.org). Our findings show that Apo E3 is more prevalent than other isoforms. Similar findings were also reported by using the restriction analysis [10]. The prevalence of isoform E3 in six families out of seven families indicates that this is the most prevalent genotype in the selected families of Pakistan.
The four genotypes detected were ε3/ε3 (73.9%), ε2/ε3 (6.5%), ε3/ε4 (17.4%) and ε2/ε4 (2.2%). Similar results with matching frequencies of genotypes were reported in one hundred and sixty (160) Lebanese applying the identical molecular methods used here in this study: ε3/ε3 (68.75%), ε3/ε4 (16.25%), ε2/ε3 (13.75%), ε2/ε4 (0.625%), and ε4/ε4 (0.625%) [11]. Another study from Lebanese Republic revealed in 1999 determining the subsequent composition frequencies in a 155 healthy adults ε2/ε3 (12.90%), ε3/ε4 (8.39%), ε3/ε3 (77.42%) and ε2/ε4 (1.29%) [12]. Homozygous ε2/ε2 and ε4/ε4 weren't detected. Absence of homozgousity may be due to low frequency of polymorphism in Lebanese population or low sample size. Yet, Mahfouz and his colleagues found just single case of ε4/ε4 but no case of ε2/ε2 genotype from total subjected Lebanese samples (160), while Almawi and his coworkers failed to notice any of these homozygous genotypes in the 155 healthy subjects investigated [11,12].
References
- Nordestgaard BG, Chapman MJ, Humphries SE, Ginsberg HN, Masana L, Familial hypercholesterolaemia is underdiagnosed and undertreated in the general population: guidance for clinicians to prevent coronary heart disease Consensus Statement of the European Atherosclerosis Society. European heart journal, (2013).
- Goldstein JL, Brown MS. The LDL receptor. Arteriosclerosis, thrombosis, and vascular biology, (2009); 29(4): 431-438.
- Fahed AC, Nemer GM. Familial hypercholesterolemia: the lipids or the genes? Nutrition & Metabolism, (2011); 8(1): 23.
- Marks D, Thorogood M, Neil HAW, Humphries SE. A review on the diagnosis, natural history, and treatment of familial hypercholesterolaemia. Atherosclerosis, (2003); 168(1): 1-14.
- Hopkins PN. Familial hypercholesterolemia—improving treatment and meeting guidelines. International Journal of Cardiology, (2003); 89(1): 13-23.
- Hobbs HH, Brown MS, Goldstein JL. Molecular genetics of the LDL receptor gene in familial hypercholesterolemia. Human Mutation, (1992); 1(6): 445-466.
- Ahmad M, Afzal S, Malik IA, Mushtaq S, Mubarik A. An autopsy study of sudden cardiac death. JOURNAL-PAKISTAN MEDICAL ASSOCIATION, (2005); 55(4): 149.
- Sambrook J, Russell DW. Rapid isolation of mammalian DNA. Cold Spring Harbor Protocols, (2006); 2006(1): pdb. prot3514.
- Rozen S, Skaletsky H (1999) Primer3 on the WWW for general users and for biologist programmers. Bioinformatics methods and protocols: Springer. pp. 365-386.
- Seet WT, Mary Anne TJA, Yen TS. Apolipoprotein E genotyping in the Malay, Chinese and Indian ethnic groups in Malaysia—a study on the distribution of the different apoE alleles and genotypes. Clinica Chimica Acta, (2004); 340(1): 201-205.
- Mahfouz RA, Sabbagh AS, Zahed LF, Mahfoud ZR, Kalmoni RF, et al. Apolipoprotein E gene polymorphism and allele frequencies in the Lebanese population. Molecular Biology Reports, (2006); 33(2): 145-149.
- Almawi W, Hassan G, Habbal MZ. Apolipoprotein E polymorphism in a healthy Lebanese population. Medical Principles and Practice, (1999); 8(2): 105-110.