Full Length Research Article
In vivo investigation of interactions between replisome components in Escherichia coli: An expanded model for the processivity switch
Atif A. Patoli*1,2, Bushra B. Patoli1,2
Adv. life sci., vol. 7, no. 2, pp. 66-71, February 2020
*- Corresponding Author: Atif A. Patoli (Email: atifpatoli@gmail.com)
Authors' Affiliations
2. Institute of Microbiology, University of Sindh, Jamshoro – Pakistan
Abstract![]()
Introduction
Methods
Results
Discussion
References
Abstract
Background: Protein interactions within the replisome (a highly coordinated protein complex) are crucial to maintain temporal and spatial regulation for high fidelity DNA synthesis in Escherichia coli (E. coli). A key component of these interactions is the processivity switch, ensuring smooth transition of the replicative DNA polymerase III (Pol III) between Okazaki fragments on the lagging strand. Multiple interaction studies between replisome components have been performed to indicate the essential roles of Pol III (DnaE), β-clamp, DnaB helicase, DNA and the t (DnaX) subunit for this switch.
Methods: Known interacting regions of both DnaE and various truncated versions of t were chosen for co-expression in E. coli. Differences in the growth pattern of cells co-expressing various truncated versions of DnaX and DnaE, on liquid and solid media were subsequently analyzed. Based on in vivo analyses to explore the interactions between these components, an expanded model for the processivity switch is presented here.
Results: The analyses suggest that residues 481-643 of t are sufficient to establish a functional interaction with the DnaB helicase and DnaE during replication, while residues 461-480 of t interact with the C-terminal tail of DnaE to disengage Pol III from the β-clamp during processivity switching. We also propose that residues 430-460 of t are involved in sensing the DNA structure required for the processivity switch.
Conclusion: These observations expand the current understanding of processivity switching and help dissect the regions of t utilized for binding to different replisome components such as DnaB helicase, polymerase and DNA.
Keywords: Processivity Switch; Clamp Loader; DnaE; DnaX; DnaB Helicase
DNA replication is a highly orchestrated process to ensure the rapid and accurate replication of the DNA sequence. The majority of DNA replication in E. coli is carried out by Polymerase III (Pol III), termed the replicative polymerase [1]. The replicative polymerase interacts with other proteins and forms a large multi-subunit complex, referred to as the replisome, consisting of at least 13 subunits [2], including the replicative polymerase, β-clamp processivity factor, clamp loader complex and the DnaB DNA helicase [3-5]. Pol III is a heterotrimer of α, ε and θ subunits [6]. The actual polymerization unit of Pol III is the α subunit [7] which is encoded by the dnaE gene [8]. The DnaE protein is a 129kDa protein and constitutes 1160 amino acids. The C-terminal 243 amino acid region is involved in making direct contact with β-clamp and contains two β-binding motifs [9]. The first motif is located at residues 920–924, while the second is located near the extreme C-terminus of the full-length protein at residues 1154–1159 [10]. These motifs are generally referred to as the internal Clamp Binding Motif (i-CBM) and external Clamp Binding Motif (e-CBM), respectively. The β-clamp processivity factor is a homodimeric protein that forms a ring-shaped molecule that encircles DNA. The clamp loader complex is composed of five different subunits i.e. γ3, δ1, δ’1, χ1, ψ1[7]. In E. coli the holoenzyme contains two t subunits and one γ subunit [11]. Both γ and t are encoded by the same gene (dnaX), but γ lacks the 24 kDa C-terminal region, due to a ribosomal frame shift in the dnaX gene [12, 13]. Interactions between the multiple protein components of the replisome are crucial in maintaining both spatial and temporal regulation of the process. One key stage in the process is the “processivity switch”, where the lagging strand DNA polymerase jumps from the junction of the completed Okazaki fragment to a new primer. A model describing this recycling process is referred to as the ‘collision release’ model where the Pol III holoenzyme collides with the 5’ terminus of a downstream Okazaki fragment [14, 15]. The mechanism by which the polymerase acquires the knowledge that replication was complete, and then dissociates from β-clamp and DNA, is based on an internal competition reaction between replisome subunits. Besides β-clamp, Pol III and DNA, the t subunit of clamp loader takes part in this competition reaction [16]. The competition is modulated by DNA structure. On primed DNA, the polymerase binds to β-clamp for processive synthesis, while on completed duplex DNA the polymerase loses its affinity for β-clamp and binds to t [17]. Studies have suggested that the actual trigger for this switch is contained in the t –subunit [16].
The t -subunit in Escherichia coli organizes the Pol III holoenzyme [18]. The full-length t subunit is a 71 kDa protein containing 643 amino acid residues [19]. t has a five-domain structure [20] the N-terminal Domains I–III being identical to γ. The unique 24 kDa C-terminal fragment comprising most of Domain IV and all of Domain V (often referred to as tc; residues 430–643) is connected to Domain III by a Proline-rich tether [21]. The 8 kDa Domain IVa, (residues 430– 498) is responsible for binding to the DnaB helicase [20] and is known to bind DNA [21], but exact regions for interaction have not been reported. Using surface plasmon resonance (SPR), the 16 kDa Domain V (residues 496–643) has been shown to bind full length DnaE [20]. Jergic and others reported that the C-terminal 18 residues of Domain V of t are unstructured and are involved in making a contact with full length DnaE [21]. The cryo-EM structure of Pol-IIIα, β-clamp, exonuclease and t-subunit reported by Rafael Fernandez-Leiro and others (2015) reveals the interaction of residues 530-535 and 562-566 of domain V of t to DnaE in the absence of DNA (Figure 1) while the C-terminal 18 residues of Domain V of t could not be molded in their structure [22]. Furthermore, it has been shown that the over-expression in E. coli of Domain V of t is toxic; presumably because the over-expressed protein binds native DnaE and therefore adversely affects DNA replication [21]. The C-terminal 20 residues of DnaE (CT20) have been shown to bind tc using size exclusion chromatography (Figure 1) [17] but a specific region for CT20-binding on tc has not yet been defined. Interactions beyond the C-terminal tail of DnaE have recently been characterized. The cryo-EM structure involving various replisome components shows DnaE- t interaction utilizing 657–667 residues of DnaE (Figure 1) [21].
For biochemical and structural characterization of the interactions between DnaE and t, the broadly known interacting regions of both were chosen for initial over-expression in E. coli. Given the fact that even small scale expression of Domain V is toxic to cells [21] we chose a co-expression strategy based on the rationale that if the two over-expressed proteins interacted within the expression host this would lessen any detrimental impact on the host replication system. The co-expression relieved the toxic effect of Domain V on the expression strain, however smaller, sick colonies were observed compared to those expressing tc residues 430–643 alone. We realized that this co-expression system, affecting growth rate and colony size with different DnaX constructs, could be used as a reporter assay for in vivo analysis of the role of the 24kDa C-terminal region of the t -subunit. To dissect the requirements for DNA and DnaB helicase binding capabilities, we also included two more versions of tc truncated at residues 461 and 480. Differences in the growth pattern of cells co-expressing various truncated versions of DnaX and DnaE, on liquid and solid media were subsequently analyzed. We now propose an updated model for processivity switching in E. coli based on this analysis.
Construction of truncated versions of DnaX and DnaE
For this study, DnaE (truncated at residues ED701) and four versions of DnaX (truncated at residues XD430, XD461, XD480 and XD506) were amplified using pAP9-X-FL and E. coli K12 genomic DNA as template DNA, respectively (Figure 1). In order to achieve various combinations of t with dnaE for co-expression, the dnaE construct was inserted into the MCS-I restriction site of the pET-Duet-1 expression vector (Novagen) using NcoI and BamHI restriction sites, and the t constructs were inserted into MCS-II of the same plasmids using the NdeI and XhoI restriction sites. The expression plasmids used in this study are detailed in Table 1. DH5α cells were used for cloning and amplification of these plasmids. The presence of original wild type sequences of inserts in appropriate orientations was confirmed by DNA sequencing.
Assessment of the growth pattern in liquid and solid media
For expression purposes competent E. coli B834 (DE3) expression strains were transformed separately with these plasmids and plated on LB agar plates containing ampicillin as selection antibiotics. Fresh transformants were grown in 5 ml LB broth containing appropriate antibiotics to exponential phase (OD 650 nm ~0.6-0.8). These cultures were then used for the analysis of growth pattern on solid agar and for the generation of growth curves.
In order to assess growth on solid media a series of 10-fold dilutions were made to 10-5 in LB broth. Triplicate 10 μl samples of each dilution were then spotted onto LB agar plates containing appropriate antibiotic and 0.1 mM IPTG to induce expression of the t and dnaE constructs. Control plates without IPTG were also set up. The plates were then left to dry. Once the spots had dried the plates were incubated at 37ºC overnight.
To assess growth in liquid media, 100 μl of each culture was used to inoculate 5 ml LB broth containing appropriate antibiotic and 0.1 mM IPTG. The experiments were repeated in triplicate several times and the growth curves shown were generated from the mean values of representative triplicate sets. Control tubes without IPTG were also set up. The tubes were then incubated in a shaking water bath (180 rpm) at 37 °C. OD at 650 nm was measured hourly and mean values of periodically recorded ODs were used to generate growth curves. Two-way ANOVA and Sidak’s multiple comparisons test (GraphPad Prism v7.02) was used to analyze bacterial growth curves.
Comparative analysis of cells co-expressing various truncated versions of t with DnaEΔ701h
Domain IVa, (residues 430–498 of t) is known to bind to the DnaB helicase and additionally possesses all the determinants to bind DNA [22]. Due to lysine and arginine residues spread throughout, this domain has an overall positive nature. Bioinformatics analysis suggested several of these are conserved in related bacterial t proteins and distributed throughout the domain. Therefore, to dissect the requirements for DNA and DnaB helicase binding, Domain IV in tc was truncated randomly in two positions (i.e. residues 461 and 480). The growth of cells co-overexpressing DnaEΔ701h with DnaXΔ430, DnaXΔ480 or DnaXΔ506 were compared (Figure 2). The generation times for cells were calculated using a first order reaction equation for doubling time. The cells co-expressing DnaE701 with DnaX430 grew 89% faster than those co-expressing DnaEΔ701h with DnaXΔ506 while the cells co-expressing DnaEΔ701h with DnaXΔ480 grew 65% faster than those co-expressing DnaEΔ701h with DnaXΔ506. The two-way ANOVA was used to show p ˂ 0.0001 at all-time points after 60 minutes when induced with IPTG and 120 minutes when un-induced for cells co-expressing DnaE701 with DnaX430 and/or DnaX506. Cells co-expressing DnaEΔ701h with DnaXΔ480 had reduced viability on solid media compared with cells co-expressing DnaEΔ701h with DnaXΔ506 or DnaXΔ430 (Figure 2). Various attempts to transform the expression strain B834 (DE3) with a plasmid (pAP12-XΔ461/EΔ701h) encoding DnaXΔ461 in MCS II and DnaEΔ701h in MCS I of pET-Duet-1 failed, suggesting that this combination is toxic to the cells. From the present study, few interesting models are anticipated and discussed below (Figures 3-5).
Figures & Tables
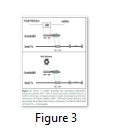
Residues 480-643 of t may establish a functional interaction with DnaB helicase and DnaE during replication
The interaction of the Pol III holoenzyme with the DnaB helicase is central to replication. Of all the subunits of the holoenzyme only t is known to exhibit interaction with DnaB [23] and that interaction is required to mediate replication fork progression. Gao and others showed that out of five domains it is Domain IV of t, that interacts with DnaB [20]. It is further known that more than one t subunits bind the helicase, suggesting that both leading and lagging strand polymerases are attached to the helicase at the same time in the replisome. Since the polymerase has to attach to the β-clamp directly at the time of active DNA synthesis, while making an indirect bridge with helicase via t, any version of t that is capable of maintaining a bridge between the polymerase and DnaB helicase can be expected to perform that role. In fact tc, the truncated version of t comprising only Domains IV and V, is capable of establishing a bridge between the polymerase and helicase and has previously been shown to restore the normal rate of replication fork progression [24]. The interaction of tc with the C-terminal of DnaE is known to function in the processivity switch [17]. Previously two functions for Domain IVa have been described, i.e. binding to DnaB helicase and binding to DNA to elicit the processivity switch. It is possible that the extra 25 N-terminal residues in DnaXΔ480 which are absent from DnaX506 may be involved in this binding of t to either DNA or DnaB helicase (Figure 3). The cells co-expressing DnaXΔ480 with DnaEΔ701h produced healthy normal colonies on solid agar as compared with cells co-expressing DnaXΔ506 with DnaEΔ701h. One possible explanation is that this version of t (DnaX Δ480) is capable interacting with the DnaB helicase. On the contrary, if these extra 25 N-terminal residues in DnaXΔ480 relative to DnaXΔ506 were required solely for DNA recognition the cells would have behaved similarly to those co-expressing DnaXΔ506 with DnaEΔ701h (i.e small colonies) as DNA recognition would be transient and therefore unlikely to retrieve the toxic effects of over-expression.Given the fact that Domain IVa is known to bind DnaB helicase, and tc is able to restore the normal rate of replication fork progression [24], it is plausible that the N-terminal 25 residues in DnaXΔ480 may similarly restore function by interaction with DnaB helicase. The normal size of the colonies co-expressing DnaXΔ480 with DnaEΔ701h indicates that residues 480 to 643 of t might be sufficient to establish a bridge between the polymerase and DnaB helicase. Holding the native polymerase via Domain V, DnaXΔ480 may bind to DnaB helicase with its N-terminal 25 residues and thus form part of the replisome but not the clamp loader complex (Figure 3). Without a sustained DnaB-DnaX480 interaction the DnaX construct could not function as part of the replisome and therefore its over-expression would be expected to be toxic as with DnaX506.
Intriguingly, DnaX480 does not show similar growth patterns to DnaX430 on solid agar and presumably differences in the two constructs are highlighted by the differing growth types. A possible explanation lies in the different modes of replication on the leading and lagging strand: DnaX480 lacks a significant portion of Domain IV and these results suggest it has some defect, potentially in the DNA binding events that coordinates the processivity switch. Such a defect could plausibly have a more significant effect on the lagging strand polymerase than on the leading strand as reflected by reduced viability observed on solid agar in comparison with the cells co-expressing DnaE701 with either DnaX430 or DnaX506 (Figure 2).
Residues 461 – 480 of t may mediate binding to the external clamp binding motif of the α-subunit during the processivity switch.
A dynamic and functional replisome requires stable interactions between the clamp loader, DnaB helicase, polymerase and β-clamp, while maintaining the property of switching the polymerase “on” and “off” on β-clamp during lagging strand synthesis. It has been established that the C-terminal region of the α-subunit is crucial to this switch (generally termed the processivity switch) as this region switches between binding β-clamp and t subunit of the clamp loader [17]. Our model suggests that DnaXΔ480 might be sufficient to hold the polymerase at the replication fork through contacts with DnaB helicase. Multiple attempts to transform the expression strain B834 (DE3) with a plasmid (pAP12-XΔ461/EΔ701h) encoding DnaXΔ461 from MCS II and DnaEΔ701h from MCS I of pET-Duet-1 failed, suggesting that even very low levels of expression of DnaXΔ461 from the tightly controlled pETDuet-1, is toxic to the cells. This toxic effect of DnaXΔ461 is presumably due to this construct possessing an additional 19 residues at its N-terminal end compared with DnaXΔ480. The additional 19 residues potentially enhance the binding affinity of DnaXΔ461 for one or more replisome components. Previously the C-terminal 20 residues of DnaE containing e-CBM (residues 1154–1159 [10] have been shown to bind tc [15]. Since a specific region for tc making contact with CT20 residues of DnaE has not yet been defined, we postulate that residues 461-480 block the e-CBM of DnaE to prevent the polymerase-clamp binding required for processive synthesis (Figure 4). A homologous interaction is seen in the crystal structure of Thermus aquaticus tc and α [25] where NTD of Taq tc (corresponding to domain IV of tc of E. coli) is seen to bind β-binding domain of Pol IIIα. We assume that DnaXΔ461 sequesters the native polymerases and DnaB helicase, therefore its over-expression has the potential to be toxic. Further studies are required to extend these observations.
Residues 430-480 appear to be required for appropriate regulation and function of tc
Lopez de Saro and others showed that in addition to polymerase and β-clamp, the t subunit is needed for the processivity switch [17]. They demonstrated that the tc section of t was sufficient to execute this switch and also showed that tc can bind DNA and sense the difference in structure between primed DNA and duplex DNA, coupling this DNA sensing to separation of polymerase from the β-clamp. The DNA-sensing region was shown to be contained in Domain IVa of t [20]. In the present study, we have postulated that DnaXΔ480 might be sufficient to hold the polymerase at the replication fork through contacts with DnaB helicase, while residues 461-480 of t could block the e-CBM of DnaE during processivity switching. Cells over-expressing DnaXΔ430 with/without DnaEΔ701h behaved as normal, suggesting that tc is sufficient for executing the processivity switch; in fact tc has already been shown to restore the normal rate of replication fork progression [24]. Since DnaXΔ480 appears to partially function in the replisome and presumably binds DnaB but shows some growth defects, this would suggest that the DNA-sensing region may be localized in residues 430-480. Once again further studies are required to extend the observations beyond this in vivo analysis.
Conclusion and future directions: A possible model for various interactions of t with DNA, DnaB helicase, DnaE polymerase is shown in Figure 5. The model presented extends the current knowledge of processivity switch and provides a solid basis for studies going forward. Site-directed mutation studies of the positively charged regions in Domain IVa, for example, would deepen understanding of the residues involved with DNA binding/sensing function. Purification of the various DnaX truncations for in vitro analysis by Surface Plasmon Resonance or Isothermal Calorimetry with DnaB and DnaE constructs could also be employed to further probe the processivity switch and provide additional details to map the interactions of this key component of the bacterial replisome.
Acknowledgement
We would like to thank HEC Pakistan and University of Sindh, Jamshoro for funding. We pay special regards to Dr. Karen A. Bunting and Dr. Jody A. Winter for their guidance during this research work.
Authors' Contribution
A.A. Patoli designed the project. Cloning of various constructs i.e. experimental work was done by A.A. Patoli and B. B. Patoli. Both the authors contributed in writing the manuscript.
Authors declare that the current research work does not involve any human participant or animal model. The project was funded by HEC Pakistan and University of Sindh, Jamshoro, and executed in the School of Biology, Queens Medical Centre, University of Nottingham, Nottingham, NG7 2UH, UK. It is also declared herewith that there is no conflict of interests in the current manuscript.
- Kim DR, McHenry CS. Identification of the beta-binding domain of the alpha subunit of Escherichia coli polymerase III holoenzyme. Journal of Biological Chemistry, (1996); 271(34): 20699-20704.
- Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Advances in Protein Chemistry (2005); 71401-440.
- Robinson A, van Oijen AM. Bacterial replication, transcription and translation: mechanistic insights from single-molecule biochemical studies. Nature Reviews Microbiology, (2013); 11(5): 303-315.
- Lewis JS, Spenkelink LM, Jergic S, Wood EA, Monachino E, et al. Single-molecule visualization of fast polymerase turnover in the bacterial replisome. Elife, (2017); 6.
- Lewis JS, Jergic S, Dixon NE. The E. coli DNA Replication Fork. Enzymes, (2016); 3931-88.
- McHenry CS, Crow W. DNA polymerase III of Escherichia coli. Purification and identification of subunits. Journal of Biological Chemistry, (1979); 254(5): 1748-1753.
- Johnson A, O'Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annual Review of Biochemistry, (2005); 74283-315.
- Welch MM, McHenry CS. Cloning and identification of the product of the dnaE gene of Escherichia coli. Journal of Bacteriology, (1982); 152(1): 351-356.
- Lamers MH, Georgescu RE, Lee SG, O'Donnell M, Kuriyan J. Crystal structure of the catalytic alpha subunit of E. coli replicative DNA polymerase III. Cell, (2006); 126(5): 881-892.
- Dohrmann PR, McHenry CS. A bipartite polymerase-processivity factor interaction: only the internal beta binding site of the alpha subunit is required for processive replication by the DNA polymerase III holoenzyme. Journal of Molecular Biology, (2005); 350(2): 228-239.
- Dohrmann PR, Correa R, Frisch RL, Rosenberg SM, McHenry CS. The DNA polymerase III holoenzyme contains gamma and is not a trimeric polymerase. Nucleic Acids Research, (2016); 44(3): 1285-1297.
- Flower AM, McHenry CS. The gamma subunit of DNA polymerase III holoenzyme of Escherichia coli is produced by ribosomal frameshifting. Proceedings of the National Academy of Sciences of the United States of America, (1990); 87(10): 3713-3717.
- Yuan Q, Dohrmann PR, Sutton MD, McHenry CS. DNA Polymerase III, but Not Polymerase IV, Must Be Bound to a t-Containing DnaX Complex to Enable Exchange into Replication Forks. The Journal of Biological Chemistry, (2016); 291(22): 11727-11735.
- Georgescu RE, Kurth I, Yao NY, Stewart J, Yurieva O, et al. Mechanism of polymerase collision release from sliding clamps on the lagging strand. EMBO Journal, (2009); 28(19): 2981-2991.
- Dohrmann PR, Manhart CM, Downey CD, McHenry CS. The rate of polymerase release upon filling the gap between Okazaki fragments is inadequate to support cycling during lagging strand synthesis. Journal of Molecular Biology, (2011); 414(1): 15-27.
- Leu FP, Georgescu R, O'Donnell M. Mechanism of the E. coli tau processivity switch during lagging-strand synthesis. Molecular Celll, (2003); 11(2): 315-327.
- Lopez de Saro FJ, Georgescu RE, O'Donnell M. A peptide switch regulates DNA polymerase processivity. Proceedings of the National Academy of Sciences USA, (2003); 100(25): 14689-14694.
- O'Donnell M. Replisome architecture and dynamics in Escherichia coli. Journal of Biological Chemistry, (2006); 281(16): 10653-10656.
- Su XC, Jergic S, Keniry MA, Dixon NE, Otting G. Solution structure of Domains IVa and V of the tau subunit of Escherichia coli DNA polymerase III and interaction with the alpha subunit. Nucleic Acids Research, (2007); 35(9): 2825-2832.
- Gao D, McHenry CS. t Binds and Organizes Escherichia coli Replication Proteins through Distinct Domains: PARTIAL PROTEOLYSIS OF TERMINALLY TAGGED t TO DETERMINE CANDIDATE DOMAINS AND TO ASSIGN DOMAIN V AS THE α BINDING DOMAIN. Journal of Biological Chemistry, (2001); 276(6): 4433-4440.
- Jergic S, Ozawa K, Williams NK, Su XC, Scott DD, et al. The unstructured C-terminus of the tau subunit of Escherichia coli DNA polymerase III holoenzyme is the site of interaction with the alpha subunit. Nucleic Acids Research, (2007); 35(9): 2813-2824.
- Fernandez-Leiro R, Conrad J, Scheres SH, Lamers MH. cryo-EM structures of the E. coli replicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease and tau. Elife, (2015); 4.
- Kim S, Dallmann HG, McHenry CS, Marians KJ. Coupling of a replicative polymerase and helicase: a tau-DnaB interaction mediates rapid replication fork movement. Cell, (1996); 84(4): 643-650.
- Dallmann HG, Kim S, Pritchard AE, Marians KJ, McHenry CS. Characterization of the Unique C Terminus of theEscherichia coli t DnaX Protein: MONOMERIC C-t BINDS α AND DnaB AND CAN PARTIALLY REPLACE t IN RECONSTITUTED REPLICATION FORKS. Journal of Biological Chemistry, (2000); 275(20): 15512-15519.
- Liu B, Lin J, Steitz TA. Structure of the PolIIIalpha-tauc-DNA complex suggests an atomic model of the replisome. Structure, (2013); 21(4): 658-664.
This work is licensed under a Creative Commons Attribution-Non Commercial 4.0 International License. To read the copy of this license please visit: https://creativecommons.org/licenses/by-nc/4.0