![]()
Characterization of mercury resistant and growth promoting Enterobacter sp. from rhizosphere to use as a biofertilizer
Nageena Mobeen, Zakia Latif*
Adv. life sci., vol. 3, no. 2, pp. 36-41, February 2016
*- Corresponding Author: Zakia Latif (Email: zakia.mmg@pu.edu.pk)
Authors' Affiliation
Abstract![]()
Introduction
Methods
Results
Discussion
References
Abstract
Background: Mercury occurs naturally in environment, it is heavy metal that exists in three chemical forms like elemental mercury, inorganic mercury and organic mercury. All forms of mercury are problematic for living organism. Currently, the contamination of agricultural land and water systems with mercury has become one of the major environmental issues. The cheapest mode to remove mercury metal and its other forms from the ecosystem is the use of microorganisms.
Methods: In this study, initially bacterial species were isolated and purified from nodule like structures on roots and stems of plants on MacConkey agar medium. Further screening for resistance to mercury was done on N- agar medium supplemented with different concentration of HgCl2 (20, 30, 40 and 50 µg/mL). Well plate method was used for the determination of bacterial strains having maximum ability to detoxify mercury. Selected bacterial strains were subjected to different biochemical tests for characterization and other metabolic tests were also performed to characterize their capabilities.
Results: All strains were highly resistant to HgCl2 at the concentration of 20 µg/mL and moderately resistant at 30 µg/mL. Bacterial strain S-2 was moderately resistant and S-3 was least resistant at 40 µg/ml whereas S-2 was least resistant at 50 µg/ml. Selected bacterial strains were positive for nitrogen fixation and protease production, negative for phosphate solubilization but only S-1 was positive for hydrogen cyanide (HCN). Bacterial species were molecular characterized by 16S rDNA sequencing as Enterobacter cloacae (KJ857483, KJ857484 and KJ857485: NCBI GeneBank).
Conclusion: Selected Enterobacter sp. exhibiting multiple characteristics can be used as biofertilizer in mercury polluted land for sustainable agriculture.
Keywords: Enterobacter, Mercury, 16S rDNA, H2S, Biofertilizer
Introduction
Environmental pollution is of serious concern and interest in bacterial resistance to heavy metals is of practical significance. The release of hazardous chemicals from industrial sector has major role in polluting our environment. Hg2+, Pb2+, Cd2+ and Cu2+ usually release directly in the marine and terrestrial ecosystem. These metal ions are non-degradable and highly toxic and accumulation of such heavy metals in agricultural soils play crucial role in increasing the concentration of heavy metals in the environment. Moreover, vehicle exhaust, waste incineration and disposal also lead to the build-up of heavy metals in the agricultural soils. Thereby presenting risk to public health due to transfer of metals from soil to plants. These heavy metals were included in 2013 top ten list of the priority hazardous substances [1].
Mercury contamination is of true concern among other heavy metals, in the light of high degree of lethal effects to living organisms. Mercury is rarely present in nature and is the only metallic element that is liquid at ordinary temperature and known as quicksilver. Long term exposure to mercury vapor primarily affects the central nervous system and it also accumulates in kidney tissues, directly causing renal toxicity. High concentration of Hg2+ as it is hazardous to living health causes impairment of pulmonary function, chest pain, dyspnoea and brain disorders. Mercury discharge should be prevented from industrial sites by efficient and cost-effective end-of-pipe treatment technologies. Various types of technology are available for removal of mercury from contaminated water and wastewater but all are very expensive. Developments in the field of environment biotechnology indicate that bacteria, fungi, yeasts and algae can remove heavy metals from aqueous solution by adsorption. Organisms with metabolic activities have the ability to transform toxic contaminants to the less toxic compounds that can be integrated into our biogeochemical cycles [2].
Many research studies have reported that besides their part in securing the plants from harmful impact of metals and also as a biological vector, the plant growth promoting bacteria play an essential role in increasing the fertility of soil and annual crop production by providing vital supplements for their growth [3-5]. Enterobacter cloacae strains have been shown to colonize and benefit plant growth in various crops, such as soybean, cucumber, corn, rice and ginger. Previous studies of several plant-origin isolates have shown that E. cloacae has antagonistic effects against the oomycete pathogen, Pythium ultimum [6]; fungal pathogens, Fusarium moniliforme and Fusarium oxysporum and the bacterial pathogen, Ralstonia solanacearum. Additionally, several other strains of E. cloacae are considered to be plant growth-promoting rhizobacteria (PGPR). Enterobacter cloacae have been used as a biological control for plant disease such as the seed-rotting oomycete in Pythium ultimum and used to control insect pests on mulberry leaves and suppress disease.
The main focus of this research work was to isolate Enterobacter species exhibiting multiple characteristics such as resistant to high dose of mercury, resistant to fungal pathogens, capable of nitrogen fixation and other growth promoting substances. These strains can be used as a biofertilizer in polluted areas irrigated with mercury contaminated industrial effluents.
Methods
Collection of samples
Different explants samples such as Rose (root section), Lemon (stem section) and Mosaami (stem section) were collected from Botanical garden of University of the Punjab, Lahore Pakistan. Special care was taken to avoid contamination and stored in the refrigerator in zip-lock plastic bags.
Culture of bacteria on selective medium (MacConkey agar)
All explant samples were rinsed with tap water for removing soil and hazardous materials. Nodule like structures present on the roots and stems were picked up and surface sterilized with 95% ethanol for 1 minute. After thorough washing, surface sterilized nodules were finely homogenized in sterilized pestle and mortal to prepare 0.1% suspension in autoclaved distilled water. Two fold dilution up to 106 were prepared. From each dilution, 50 µL was spread on the MacConkey agar plates with the help of sterilized glass spreader. The plates were incubated at 30oC for 24-48 hours. Morphological different colonies were selected and re-streaked on the same medium for purification.
Screening of HgCl2 resistant and H2S producing bacterial strains
The isolated strains were qualitatively screened for their ability to resist mercury by using different concentrations of HgCl2 such as 20, 30, 40 and 50 µg/mL in well plate assay [7]. Optical density (OD600) of overnight bacterial cultures was fixed at 0.1 and 100 µL of each culture was spread on N-agar plates. Four wells (5 mm in diameter) were prepared on each N-agar plate with the help of sterilized pasture pipette. Sterilized HgCl2 solution (20 µL) of each concentration was poured into each well separately. The plates were incubated at 30○C for 24-48 hours and observed the resistance. Selected bacterial strains were also checked for their ability to produce hydrogen sulfide (H2S) by following the method described by Amin and Latif [8].
Antibiotics resistance test
Antibiotic susceptibility test of selected bacterial strains was performed against using six different antibiotics including kanamycin, streptomycin, tetracycline, ceftriaxone (30 µg/mL each) rifampin, gentamycin and ampicillin (10 µg/mL each). Each 100 µL overnight grown bacterial culture (0.1 at OD600nm) was spread on the N-agar plates and four wells (5mm in diameter) were prepared on plate as in the case of HgCl2 resistant test. The solution of each antibiotic (20 µL) was poured into the respective well and incubated at 30°C for 24-48 hours. Antibiotic susceptibility was determined by measuring the size of inhibition zone.
Biochemical characterization
Gram reaction of all purified bacterial strains was done by gram’s staining [9]. For biochemical characterization of selected bacterial strains further tests such as Catalase (using 3% H2O2) and oxidase (using p-aminodimethylaniline oxalate) activities were also checked [9].
Metabolic fingerprinting of selected bacterial strains
i) Nitrogen fixation
Nitrogen free mannitol agar was used to observe the growth of nitrogen fixing bacteria. Nitrogen free mannitol agar [10] was prepared and poured in autoclaved plates. Bacterial cultures were streaked on plates. Plates were sealed carefully with the para-film and incubated at 28°C for 24-72 hrs. After incubation, appearance of growth on culture plate was observed and recorded.
ii) Phosphate solubilization
Phosphate solubilization test was performed to observe the ability of bacterial strains to solubilize the phosphate by producing halo zones on Pikovskaya’s agar medium [11] supplemented with 5 g/L tricalcium phosphate. Cultures were incubated at 28°C for 2-3 days and results were observed for clear zones of phosphate solubilization.
iii) Hydrogen cyanide production (HCN)
Hydrogen cyanide (HCN) activity of bacterial culture was determined as described by [10]. Bacterial strains were cultured on N-agar supplemented with glycine (4.4 g/l). Whatmann filter paper NO.1 soaked in a solution of 2% sodium carbonate in 0.5% Picric acid was placed at the top of the culture plate. Plates were sealed with parafilm in order to avoid the escape of gaseous metabolite (HCN). Plates were incubated at 28○C for 5 days and observe the color change on the filter paper.
iv) Protease production
To determine the ability of strains for the production of protease enzyme, each bacterial strain from the overnight grown culture on N-agar was spotted onto skim milk agar containing skimmed milk 28.0, casein enzymatic hydrolate 5.0, yeast extract 2.0, dextrose 1.0 and agar 15.0 g/L [12] plates and incubated at 37ºC for 48 hours. After incubation, plates were observed for the appearance of colorless clear zones around each bacterial colony.
16S rDNA sequencing
Three strains showing promising results were selected for ribotyping. 16S rDNA sequencing was undertaken and sequenced by Macrogen sequencing facility at Korea. The sequences obtained were BLAST on NCBI against already reported nucleotide sequence and were also submitted to gene bank for accession numbers.
Phylogenetic analysis of selected nitrogen fixing bacterial isolates
Phylogenetic analysis of selected isolates was performed by multiple sequence alignment with different bacterial species through neighbor joining method using MEGA 4 software with Bootstrapping value 1000 (No. of data sets).
Results
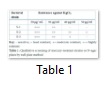
Total eighteen bacterial strains were isolated on MacConkey agar medium from three different plant sources and only three bacterial strains showed resistant against 30, 40 and 50 µg/mL respectively (Table 1) by using well plate method. It is reported that microorganisms resistant to mercury (Fig.1) are hydrogen sulfide (H2S) producers which are supposed to be involved in the transformation of toxic form of mercury (Hg+2) by reacting with H2S into nontoxic form HgS [8, 13]. Same was confirmed qualitatively by growing selected strains on lead acetate medium. So, three strains selected on the basis of mercury resistant were appeared as black color colonies.
Antibiotic resistance of selected strains was performed against different antibiotics by observing the zone of inhibition in well plate method. It was observed that all strains were resistant against kanamycin, rifampin, gentamycin, streptomycin, and ampicillin but were sensitive to tetracycline and ceftriaxone.
Molecular characterization of the selected bacterial strains was done by 16S rDNA sequencing. The BLAST query revealed that S-1, S-2 and S-3 16S rRNA gene were homologous to Enterobacter cloacae. Other close matches to Enterobacter cloacae KJ857485 included Enterobacter cloacae NR118011 (71% similarity). Likewise, the gene of Enterobacter cloacae KJ857483 and Enterobacter cloacae KJ857484 was similar to Enterobacter cloacae NR117679 and E. cloacae NR102794 (65% similarity), The bacterial isolates also show similarity (16S rRNA gene) among themselves. The maximum similarity was found between Enterobacter cloacae KJ857485 and E. cloacae KJ857483 as well as E. cloacae KJ857484 (100% similarity). Phylogenetic tree based on 16S rRNA gene sequence comparison shows the relationship between members of genera Enterobacter. Molecular characterization of the selected bacterial strains was done by 16S rDNA sequencing. The BLAST query revealed that S-1, S-2 and S-3 16S rRNA gene were homologous to Enterobacter cloacae. Other close matches to Enterobacter cloacae KJ857485 included Enterobacter cloacae NR118011 (71% similarity). Likewise, the gene of Enterobacter cloacae KJ857483 and Enterobacter cloacae KJ857484 was similar to Enterobacter cloacae NR117679 and E. cloacae NR102794 (65% similarity), The bacterial isolates also show similarity (16S rRNA gene) among themselves.
The maximum similarity was found between Enterobacter cloacae KJ857485 and E. cloacae KJ857483 as well as E. cloacae KJ857484 (100% similarity). Phylogenetic tree based on 16S rRNA gene sequence comparison shows the relationship between members of genera Enterobacter.
Data and Tables
Discussion
Heavy metals occur naturally in our environment and are constantly contaminating our soil and water sources through different anthropogenic activities. Industrial waste contains heavy metals and other organic pollutants possess serious damage to our environment. Among heavy metal contamination, mercury contamination is of great importance and is among the most alarming issue in the world. Throughout the world concentration of mercury increases day by day that is an alarming situation. Nature has developed a system in some microorganisms that they have potential to remediate heavy metals from the environment.
The present study was designed to search out mercury resistant Enterobacter sp. from roots of different plants for the remediation of mercury and as plant growth promoter in polluted agriculture land. It is in common practice in developing countries that industrial waste is discharged in agriculture land without any proper treatment to remove the dangerous heavy metals. It is reported that mercury resistant bacteria could remove mercury from contaminated areas [8, 14, 15]. Keeping in mind the above alarming scenario, different bacterial species were isolated on MacConkey agar medium. Mercury and drug resistant of microorganism was checked by simple well plate method. The basic reason to adopt this method was its simplicity and cost-effectiveness. Their ability to resist mercury was determined against different concentrations (20, 30, 40, 50 µg/mL) of HgCl2.
Antibiotic resistance of strains was performed against different antibiotics by observing the zone of inhibition in well plate method. It has been reported in the literature that Enterobacter species were sensitive to tetracycline and ceftriaxone drug. These drugs could be used as drug of choice against them [16]. Selected gram negative bacterial strains were catalase and protease positive but negative for oxidase and phosphate tests. One bacterial strain S-1 was positive for hydrogen cyanide test. Selected bacterial strains were also found because nitrogen fixer as they have the ability to grow on Nitrogen-free Mannitol agar medium. Nitrogen is generally considered one of the major limiting nutrients in plant growth. The biological process responsible for reduction of molecular nitrogen into ammonia is referred as nitrogen fixation by the aid of nitrogenase enzyme. A wide variety of bacteria are capable of fixing nitrogen. In this way, they increase soil fertility and enhance plant growth promotion. In many research studies the ability of bacteria to fix nitrogen has been reported [17].
It has been previously reported that Enterobacter species EMB21 has the potential to remediate mercury more efficiently [18]. In the literature it is also reported that Enterobacter sakazaki has the ability to fix nitrogen [19]. Our studies exactly match with the above reported finding. The present study is first report that selected Enterobacter cloacea exhibited dual characteristics. The selected species can be used not only to remediate mercury from the polluted environment but can be used as biofertilizer as well.
References
- Tangahu BV, Abdullah SRS, Basri H, Idris M, Anuar N, et al. Phytoremediation of wastewater containing lead (Pb) in pilot reed bed using Scirpus grossus. International Journal of Phytoremediation, (2013); 15(7): 663-676.
- Amin A, Latif Z. Phytotoxicity of Hg and its Detoxification through Microorganisms in Soil. Advancements in Life Sciences, (2015); 2(2): 98-105.
- Zaidi A, Khan S. Interactive effect of rhizotrophic microorganisms on growth, yield, and nutrient uptake of wheat. Journal of plant Nutrition, (2005); 28(12): 2079-2092.
- Kamran MA, Eqani SAMAS, Bibi S, Xu R-k, Monis MFH, et al. Bioaccumulation of nickel by E. sativa and role of plant growth promoting rhizobacteria (PGPRs) under nickel stress. Ecotoxicology and Environmental Safety, (2016); 126256-263.
- Szymańska S, Płociniczak T, Piotrowska-Seget Z, Złoch M, Ruppel S, et al. Metabolic potential and community structure of endophytic and rhizosphere bacteria associated with the roots of the halophyte Aster tripolium L. Microbiological Research, (2016); 18268-79.
- van Dijk K, Nelson EB. Fatty acid competition as a mechanism by which Enterobacter cloacae suppresses Pythium ultimum sporangium germination and damping-off. Applied and Environmental Microbiology, (2000); 66(12): 5340-5347.
- Zeroual Y, Moutaouakkil A, Blaghen M. Volatilization of mercury by immobilized bacteria (Klebsiella pneumoniae) in different support by using fluidized bed bioreactor. Current Microbiology, (2001); 43(5): 322-327.
- Amin A, Latif Z. Detoxification of mercury pollutant by immobilized yeast strain Candida xylopsoci. Pakistan Journal of Botany, (2013); 45(4): 1437-1442.
- Scherrer R. Gram's staining reaction, Gram types and cell walls of bacteria. Trends in Biochemical Sciences, (1984); 9(5): 242-245.
- Ahmad F, Ahmad I, Khan M. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiological Research, (2008); 163(2): 173-181.
- Cappuccino JG, Sherman N. In Microbiology: a laboratory manual. (2008); p 9; Pearson/Benjamin Cummings. England
- Joshi GK, Kumar S, Sharma V. Production of moderately halotolerant, SDS stable alkaline protease from Bacillus cereus MTCC 6840 isolated from lake Nainital, Uttaranchal state, India. Brazilian Journal of Microbiology, (2007); 38(4): 773-779.
- Ono BI, Kijima K, Ishii N, Kawato T, Matsuda A, et al. Regulation of sulphate assimilation in Saccharomyces cerevisiae. Yeast, (1996); 12(11): 1153-1162.
- Amin A, Latif Z. Isolation and characterization of H2S producing yeast to detoxify mercury containing compounds. International Research Journal of Microbiology, (2011); 2517-525.
- Teng Y, Wang X, Li L, Li Z, Luo Y. Rhizobia and their bio-partners as novel drivers for functional remediation in contaminated soils. Frontiers in Plant Science, (2015); 6.
- Sedgley C, Samaranayake L. Antimicrobial susceptibility of oral isolates of Enterobacter cloacae and Klebsiella pneumoniae from a southern Chinese population. Oral Microbiology and Immunology, (1998); 13(5): 315-321.
- Sy A, Giraud E, Jourand P, Garcia N, Willems A, et al. Methylotrophic Methylobacteriumbacteria nodulate and fix nitrogen in symbiosis with legumes. Journal of Bacteriology, (2001); 183(1): 214-220.
- Sinha A, Kumar S, Khare SK. Biochemical basis of mercury remediation and bioaccumulation by Enterobacter sp. EMB21. Applied Biochemistry and Biotechnology, (2013); 169(1): 256-267.
- Mehnaz S, Baig DN, Lazarovits G. Genetic and phenotypic diversity of plant growth promoting rhizobacteria isolated from sugarcane plants growing in Pakistan. Journal of Microbiology and Biotechnolology, (2010); 20(12): 1614-1623.