![]()
Detection of bacterial load in drinking water samples by 16s rRNA ribotyping and RAPD analysis
Mariyam Zameer1, Saleha Mahmood1, Zubaria Mushtaq1, Bushra Tabasum2*, Qurban Ali2, Nasir Mahmood3, Nadia Jamil1, Soniya Munir1
Adv. life sci., vol. 2, no. 3, pp. 135-141, May 2015
*Corresponding Author: Bushra Tabasum (Email: bushra.cemb@gmail.com)
Author Affiliations[Date Received: 09/02/2015; Date Revised: 20/05/2015; Date Published Online: 25/05/2015]
Abstract![]()
Introduction
Methods
Results
Discussion
References
Abstract
Background: Safe and healthy drinking water is inaccessible to more than 20% of the world population. Among some major risks to safety of potable water, contamination with pathogenic microorganisms is the most alarming and harmful Therefore, it is needed to develop and implement fast and accurate methods for the detection of bacterial contamination in water.
Methods: Biological analysis of drinking water samples obtained from nine different collection points of Lahore city was carried out and total of six different bacterial strains were isolated. Biochemical characterization was done under standard laboratory conditions. Molecular identification of these isolates was done by using random amplified polymorphic DNA (RAPD) analysis.
Results: The drinking water sample collected from Punjab University showed highest bacterial count 1066/0.5 ml of drinking water while residential area of University of the Punjab contained least number of bacterial counts i.e., 38/0.5 ml of drinking water. Amplification patterns of isolates SZ1, SZ3, SZ4 and SZ6 obtained by RAPD were found similar to genus Bacillus. While, SZ2 and SZ5 had unique amplification patterns identical to Bacillus megaterium. All the six bacterial strains were tested for the presence of protease, lipase, cellulase, and amylase. Strain SZ2 gave positive result for all of them except amylase.
Conclusion: Tube well water of Punjab University area of Lahore is safe for drinking purpose except admin block tube. It is recommended to monitor the bacteriological load of drinking water at regular intervals in order to control water borne bacterial diseases.
Key words: Drinking water, Bacteriological contamination, RAPD analysis, 16S rRNA gene amplification
Introduction
Access to safe drinking water is one of the major challenges of the 21st century [1]. About 20% of the world population does not have access to pure drinking water and the death rate associated with inadequate sanitation and impure drinking water consumption is more than 1.7 million/year [2]. In Asian and African countries, children under five years of age are mostly affected by water borne microbial diseases. In Pakistan, bacteriological contamination of drinking water is also a serious problem. Every year some major water borne gastro intestinal diseases break out due to consumption of this water [3]. The microbiological contamination of drinking water in Punjab is highest as compared to chemical and physical pollution [4]. Ground water is one of the important natural sources of drinking water and is usually considered pure for drinking purpose but several sources could be the cause of its contamination [5]. The major sources of ground water microbial contamination are seepage from the sewage system pipelines into boreholes due to cracked casings, poor water distribution supply networks and poor construction of water supply systems [6]. Excessive growth of bacteria in drinking water distribution systems can lead to bio-fouling of distribution pipes, bio-corrosion and clogging of filters [7].
Verification of microbial quality is necessary to manage the health impacts associated with the potential microbial hazards which include analysis of source water, water present in the distribution system, household water or water received after treatment [2]. In order to minimize the potential re-growth of bacteria the best approach is the addition of disinfectants in the distribution systems such as chlorine, chlorine dioxide or monochloramine [8]. Regular monitoring of microbial quantity in drinking water is also very important to control disease outbreaks [9]. Besides all these measures testing of drinking water microbiological quality plays an essential role in the monitoring of drinking water quality and assessing protection of public health [1].
In the current research, we have performed biological analysis of drinking water samples from Punjab University (Quaid-e-Azam Campus) to assess the bacterial load as the source of contamination by molecular and microbiological techniques.
Methods
Drinking water samples
Simple random sampling (SRS) technique was followed for collecting drinking water samples by using sterilized plastic sampling bottles. Randomly nine drinking water samples were collected from two locations viz. staff colonies and tube wells. Five water samples were collected from different blocks of staff colony viz. B-Block, C-Block, D-Block, E-Block and GH-Block while four tube well water samples were collected from the areas including Botanical Garden, E-Type block, Community Centre and Admin Block. The samples were stored at 4°C.
Microbiological analysis of drinking water samples
Ten sterile autoclaved LB agar solid media plates were prepared to allow growth of bacterial colonies for 24 hours at 37°C while one plate was used as negative control. LB agar medium contained 10 g tryptone, 5 g yeast extract and 5 g NaCl and 15 g agar per 1000 ml of the medium. Under aseptic conditions, 0.5 ml of each drinking water sample was dropped on the agar plates and spreaded with subsequent incubation at 37°C for 24 hours. The overnight incubated LB agar plates were observed visually under colony counter to determine following morphological parameters of bacterial colonies viz. shape, color, size and total number of colonies per sample. Average colony size was measured by setting a suitable scale.
Six morphologically distinct bacterial colonies (Table 2) viz. SZ1, SZ2, SZ3, SZ4, SZ5 and SZ6 were selected from the solid media plate and inoculated into the 10 ml LB broth and incubated for 24-72 hours in 37°C shaker for determination of the growth rate and subsequent biochemical as well as molecular analysis. Bacterial growth rate was determined by taking the optical density (OD) at 600 nm of the each bacterial strain culture after 12, 24, 48 and 72 hours by using spectrophotometer.
Biochemical analysis and Gram staining of bacterial strains
All the six bacterial strains (SZ1-SZ6) were screened in plate assays for the production of protease [10], lipase [11], cellulase [12] and amylase [13]. The level of comparative enzyme production was determined in terms of zone formation around each spot for bacterial strains (SZ1-SZ6). The gram staining of all six bacterial strains was carried out by standard procedure [14]
Bacterial genomic DNA isolation
Bacterial genomic DNA was isolated from all six bacterial strains by using GF-1 bacterial genomic DNA extraction kit, Vivantis, USA. 1.5 ml of liquid culture of each bacterial strain was used for DNA extraction. The DNA pellet of each bacterial strain was dissolved in 50 µl of deionized autoclaved water and stored at -20°C for further molecular analysis. DNA quality was determined by agarose gel electrophoresis.
Random Amplified Polymorphic DNA (RAPD) analysis
RAPD analysis of all six bacterial strains was carried out by using two different primers, P1 5’GGGTAACGCC3’ and P2 5’CCCGTCAGCA3’ in separate reaction mixtures. Each RAPD reaction mixture contained 1X PCR buffer 1.5 mM MgCl2, 2 mM dNTPs, 100 picomoles P1 and P2 primers in separate reactions each, Taq polymerase 2.5 units, genomic DNA 3 µl and total volume of the reaction mixture was adjusted to 50 µl for each bacterial strain. The cyclic conditions for RAPD were: initial denaturation: at 95°C for 2 min, denaturation: at 95°C for 30 sec, annealing at: (25°C, 30°C and 35°C) for 2 min, elongation at 72°C for 2 min, and total cycles were 40 coupled with final extension at 72°C for 7 min and final termination at 4°C for 30 min. The amplified RAPD products were resolved on 1% agarose gel.
Ribotyping
The 16S rRNA gene of SZ2 was amplified by using the primers and conditions as described earlier [15]. The amplified 1400 bp fragment of rRNA gene was retrieved by gene clean by gene clean kit, Fermentas, USA and subjected to sequencing by using primers as described earlier [15]. The sequencing results were analyzed through Basic Local Alignment Search Tool or BLAST analysis for bacterial strain characterization at species level.
Statistical analysis
Analysis of variance for microbial load for quantitative data was carried out using the method of Steel et al. [16].
Results
The evaluation of drinking water for presence of bacteria was carried out from nine different localities of University of the Punjab. The samples were first spreaded on bacterial growth media and separated into six different strains viz., SZ1-SZ6 based on morphological features. These strains were further characterized by determining their growth rates, and subjected subsequently to molecular and biochemical analysis. Significant results were found for all studied features (Table 1a, 3a, 3b, 3c, 3d).
The water sample obtained from Admin block tube well showed maximum number of bacterial colonies (1243/0.5 ml of sample) while water sample collected from tube well from E-type houses showed least number of bacterial colonies (38/0.5 ml of sample). The water samples from B-Block, D-Block and community centre tube wells had not good water quality having 301, 163 and 160 bacterial colonies respectively (Table 1).
The bacterial strain SZ1, SZ2 and SZ5 were found to be gram positive while all remaining strains were gram negative (Table 2). All six purified bacterial strains viz., SZ1-SZ6 were subjected to genomic DNA isolation by kit method. The results indicated that in case of all bacterial strains intact DNA band of ~20 kb was observed (Fig. 1).
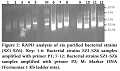
In RAPD analysis, primer 1 amplification patterns of SZ1, SZ3, SZ4 and SZ6 were found to be similar, while SZ2 and SZ5 had unique amplification patterns (Fig. 2). In order to further validate the results, the genomic DNA of all six bacterial strains was further subjected to RAPD analysis by primer 2 and same results were observed as earlier mentioned by primer 1. Bacterial strains SZ1, SZ3, SZ4 and SZ6 amplified a prominent band of 0.5 kb while strains SZ2 and SZ5 showed a prominent amplification of 0.6 kb (Fig. 2). Therefore, it is concluded that bacterial strains SZ1, SZ3, SZ4 and SZ6 genomes have not much significant genetic variation from each other and placed in the same group similarly genomes of strains SZ2 and SZ5 have genetic similarity therefore placed in second group based on RAPD analysis (Fig. 2).
All the six bacterial strains were evaluated for the production of four enzymes viz., protease, lipase, cellulase and amylase. The strain SZ2 showed overall most efficient in enzymes production except for amylase while strain SZ5 was found to be least efficient regarding all enzymes production (Table 4). Amplification of 16S rRNA gene and its subsequent sequencing was performed to characterize the bacterial strain SZ2 at specie level which was most efficient regarding enzymes production. The band of rRNA gene of 1.4 kb was amplified and purified for further sequencing reactions (Fig. 3). The BLAST analysis of sequence indicated that the sequence has absolute homology with Bacillus megaterium strain SZ2 (GenBank: JQ864207.1). It was persuasive form table 5 that the correlation among the bacterial strains of different locations was significant for admin block location with all locations except B-block and GH-block.
Data & Figure
Figure 1 Figure 2 Figure 3 Table 1 Table 1a Table 2 Table 3 Table 3a Table 3b Table 3c Table 3d Table 4 Table 5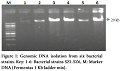











Discussion
The current study was carried out to determine the quality of drinking water collected from nine different sources inside University of the Punjab (Quaid-e-Azam campus, Lahore). The bacterial cultures were initially characterized based on their morphological features and subsequently subjected to biochemical and molecular analysis. In the current study, the water sample obtained from Admin block tube well showed maximum number of bacterial colonies (2128) while water sample collected from tube well from E-type houses showed least number of bacterial colonies viz. 38 only which showed that this drinking water has comparatively good quality for drinking purpose. The variability of bacterial load in drinking water as observed in current study was also described by other workers as well in their researches on drinking waters obtained from other geographic regions [17]. The variability in the bacterial load in drinking water is attributed to the improper cleaning of drinking water supply pipes and in sufficient chlorination of water and this finding is also in agreement an earlier study, which proposed the same fact while working on determining the quality analysis of drinking water [5].
The growth rates of six purified bacterial strains were determined for differentiation from each other regarding their generation time. The bacterial strains were divided into three categories based on growth results and these were fast growth bacteria (1-1.5 OD in first 12 hours), medium growth bacteria (0.6-0.9 OD in first 12 hours) and slow growth bacteria (0.5 OD in first 12 hours). These patterns of bacterial growth rates were observed by different workers while characterizing the novel bacterial strains isolated from different sources [18]. All six bacterial strains were screened for four enzymes viz., protease, lipase, cellulase and amylase. The strain SZ2 showed overall most efficient in enzymes production except for amylase while strain SZ5 was found to be least efficient regarding all enzymes production while remaining strains (SZ1, SZ3, SZ4 and SZ6) showed almost same efficiency regarding different enzymes production and these strains genomes also showed similarity as observed in RAPD analysis.
RAPD analysis, by using primer P1 and P2, amplified similar patterns of bacterial strains SZ1, SZ3, SZ4 and SZ6 while, SZ2 and SZ5 had unique amplification pattern. So, in RAPD analysis bacterial strains were differentiated in two separate groups as also witnessed in another study [11]. The bacterial strain SZ2 found most efficient regarding all enzymes except amylase was subjected to ribotyping by amplifying rRNA gene of 1.4 kb with subsequent direct sequencing coupled with BLAST. Sequence alignment analysis it was found to be Bacillus megaterium. As this bacterial strain is efficient in protease, lipase and cellulase production so, it can be exploited further for these enzymes production. It is suggested to screen the drinking water samples for bacteriological load at regular intervals in order to control water borne bacterial diseases.
In conclusion from this study, this can be prescribed that tube well water of Punjab University area of Lahore is safe for drinking purpose except admin block tube. It is suggested to monitor the bacteriological load of drinking water at regular intervals in order to control water borne bacterial diseases.
References
- Cabral JP. Water microbiology. Bacterial pathogens and water. International Journal of Environmental Research and Public Health, (2010); 7(10): 3657-3703.
- Ashbolt NJ. Microbial contamination of drinking water and disease outcomes in developing regions. Toxicology, (2004); 198(1): 229-238.
- Kahlown M, Tahir M, Sheikh A. Water quality status in Pakistan: Second report 2002–2003. Pakistan Council of Research in Water Resources, Islamabad, ISBN, (2004); 969-8469.
- Soomro M, Khokhar M, Hussain W, Hussain M. Drinking water Quality challenges in Pakistan. Pakistan Council of Research in Water Resources, Lahore, (2011); 17-28.
- Chitanand M, Gyananath G, Lade H. Bacterial assessment of ground water: A case study of Nanded city. Journal of Environmental Biology, (2008); 29(3): 315.
- Borah M, Dutta J, Misra AK. The bacteriological quality of drinking water in Golaghat Sub-division of Golaghat District, Assam, India. International Journal of ChemTech Research, (2010); 2(3).
- Lee SH, O'Connor JT, Banerji SK. Biologically mediated corrosion and its effects on water quality in distribution systems. Journal (American Water Works Association), (1980); 636-645.
- Der Kooij V, Lieverloo V. Distributing drinking water without disinfectant: highest achievement or height of folly? Aqua, (1999); 48(1): 31-37.
- Figueras M, Borrego JJ. New perspectives in monitoring drinking water microbial quality. International Journal of Environmental Research and Public Health, (2010); 7(12): 4179-4202.
- Zerdani I, Faid M, Malki A. Feather wastes digestion by new isolated strains Bacillus sp. in Morocco. African Journal of Biotechnology, (2004); 3(1): 67-70.
- Kumar D, Kumar L, Nagar S, Raina C, Parshad R, et al. Screening, isolation and production of lipase/esterase producing Bacillus sp. strain DVL2 and its potential evaluation in esterification and resolution reactions. Archives of Applied Science Research, (2012); 4(4): 1763-1770.
- Lynd LR, Weimer PJ, Van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiology and Molecular Biology Reviews, (2002); 66(3): 506-577.
- Deb P, Talukdar SA, Mohsina K, Sarker PK, Sayem SA. Production and partial characterization of extracellular amylase enzyme from Bacillus amyloliquefaciens P-001. Springer Plus, (2013); 2(1): 154.
- Boone DR, Castenholz RW, Garrity GM, Brenner DJ, Krieg NR, et al. Bergey's Manual® of Systematic Bacteriology. Chapter: Book Name. 2005 of publication; 2; Springer Science & Business Media.
- Niemann S, Dammann-Kalinowski T, Nagel A, Pühler A, Selbitschka W. Genetic basis of enterobacterial repetitive intergenic consensus (ERIC)-PCR fingerprint pattern in Sinorhizobium meliloti and identification of S. meliloti employing PCR primers derived from an ERIC-PCR fragment. Archives of Microbiology, (1999); 172(1): 22-30.
- Steel R, Torrie J, Dicky D Principles and Procedures of Statistics: A biometrical aproach. Chapter: Book Name. 1997 of publication; 3rd edi; 400-428. McGraw Hill Book Co. Inc.
- Seas C, Alarcon M, Aragon JC, Beneit S, Quiñonez M, et al. Surveillance of bacterial pathogens associated with acute diarrhea in Lima, Peru. International Journal of Infectious Diseases, (2000); 4(2): 96-99.
- Bain R, Bartram J, Elliott M, Matthews R, McMahan L, et al. A summary catalogue of microbial drinking water tests for low and medium resource settings. International Journal of Environmental Research and Public Health, (2012); 9(5): 1609-1625.



